Machine Learning Applications in Cancer Classification
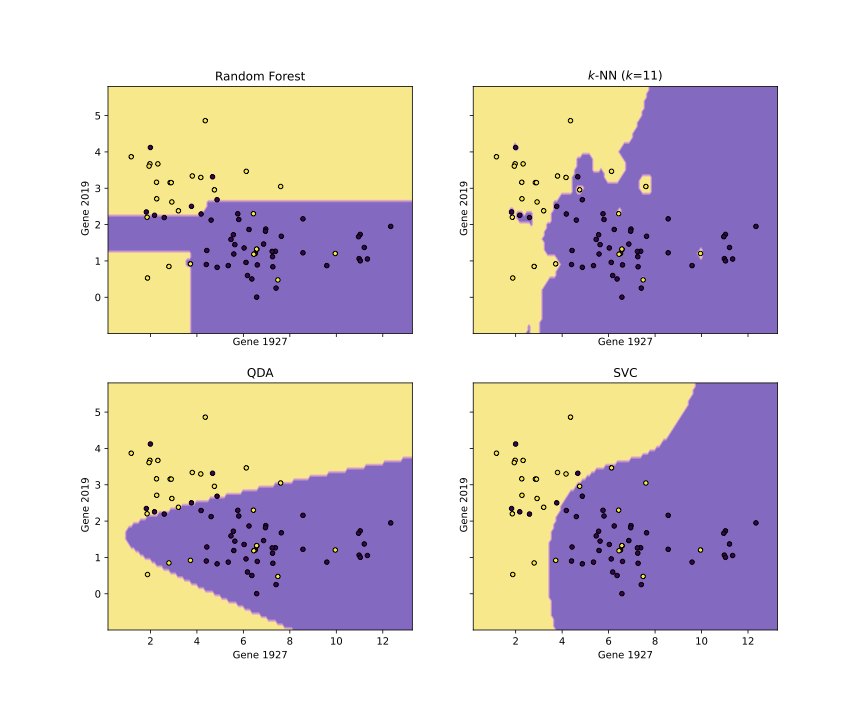
Another amazing class I took at JHU (there were a few) was Computational Molecular Medicine, a course focused on the applications of information theory and statistical learning to cancer classification. I was assigned the problem of classifying cancers as either of acute myeloid leukemia (AML) or acute lymphoblastic leukemia (ALL), using gene expression data. The project was based on research done by Golub, et al. (1999) which tackled the exact same problem. The dataset can be found on kaggle. My final report and associated code for the class is linked in the github repo above.
Reference
Golub, T R et al. (1999). Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science, 286:5439, 531-7. DOI: 10.1126/science.286.5439.531